In silico recombinant plasmid design of pHA171 with phdABCD insertion for ethidium bromide degradation
Abstract
Background: Ethidium bromide is a common reagent that is used in nucleic acid staining. However, ethidium bromide has toxic and carcinogenic properties that are harmful to the environment. Phenanthrene dioxygenase (encoded by phdA, phdB, phdC, and phdD genes) in Nocardioides sp. KP7 can oxidize the phenanthridine structure aim to eliminate carcinogenic properties.
Objective: This study aims to visualize and predict the structure, active site, and characteristics of the phenanthrene dioxygenase using bioinformatics tools.
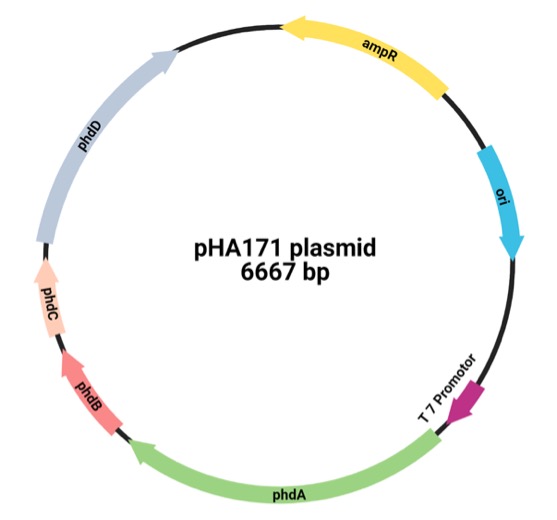
Methods: Plasmid design were prepared by inserting genes of interest phdA, phdB, phdC, and phdD from the NCBI database. Furthermore, several protein analysis tools were used for structure visualization, active site enzyme improvement, and protein characteristic of phenanthrene dioxygenase.
Results: The prediction results found that phenanthrene dioxygenase reacts with the ethidium bromide substrate through the interaction of Fe3+ ions with water. The solubility level of phenanthrene dioxygenase protein is 0.404, suggesting that the protein has low solubility. The protein isoelectric point (pI) is between 5.17 to 5.36, and the protein molecular weight is 121.143 kDa.
Conclusion: In silico analysis has supported that recombinant plasmid met characteristics for the construct which consists of gene interest and protein library.
References
Motohashi K. Development of highly sensitive and low-cost DNA agarose gel electrophoresis detection systems, and evaluation of non-mutagenic and loading dye-type DNA-staining reagents. PLoS One. 2019;14: e0222209. doi:10.1371/journal.pone.0222209
https://doi.org/10.1371/journal.pone.0222209
Saeidnia S, Abdollahi M. Are other fluorescent tags used instead of ethidium bromide safer? Daru. 2013;21: 71. doi:10.1186/2008-2231-21-71
https://doi.org/10.1186/2008-2231-21-71
Li Z, Chang P-H, Jiang W-T, Liu Y. Enhanced removal of ethidium bromide (EtBr) from aqueous solution using rectorite. J Hazard Mater. 2020;384: 121254. doi:10.1016/j.jhazmat.2019.121254
https://doi.org/10.1016/j.jhazmat.2019.121254
Youssef A, El-Ella Hussein Gad A, Ghannam H, Zedan A, Aboulthana W, Al-Sherbini A-S. Synthesis of high efficient CS/PVDC/TiO2-Au nanocomposites for photocatalytic degradation of carcinogenic ethidium bromide in sunlight. Egypt J Chem. 2020;63: 5-9. doi:10.21608/ejchem.2020.21987.2313
https://doi.org/10.21608/ejchem.2020.21987.2313
Sharma B, Dangi AK, Shukla P. Contemporary enzyme based technologies for bioremediation: A review. J Environ Manage. 2018;210: 10-22. doi:10.1016/j.jenvman.2017.12.075
https://doi.org/10.1016/j.jenvman.2017.12.075
Jia X, He Y, Jiang D, Liu C, Lu W. Construction and analysis of an engineered Escherichia coli-Pseudomonas aeruginosa co-culture consortium for phenanthrene bioremoval. Biochem Eng J. 2019;148: 214-223. doi:10.1016/j.bej.2019.05.010
https://doi.org/10.1016/j.bej.2019.05.010
Wu Y, Xu Y, Zhou N. A newly defined dioxygenase system from Mycobacterium vanbaalenii PYR-1 endowed with an enhanced activity of dihydroxylation of high-molecular-weight polyaromatic hydrocarbons. Front Environ Sci Eng. 2020;14: 14. doi:10.1007/s11783-019-1193-5
https://doi.org/10.1007/s11783-019-1193-5
Chen H, Huang R, Zhang Y-HP. Systematic comparison of co-expression of multiple recombinant thermophilic enzymes in Escherichia coli BL21(DE3). Appl Microbiol Biotechnol. 2017;101: 4481-4493. doi:10.1007/s00253-017-8206-8
https://doi.org/10.1007/s00253-017-8206-8
Vandemoortele G, Eyckerman S, Gevaert K. Pick a tag and explore the functions of your pet protein. Trends Biotechnol. 2019;37: 1078-1090. doi:10.1016/j.tibtech.2019.03.016
https://doi.org/10.1016/j.tibtech.2019.03.016
Shindo K, Ohnishi Y, Chun HK, Takahashi H, Hayashi M, Saito A, et al. Oxygenation reactions of various tricyclic fused aromatic compounds using Escherichia coli and Streptomyces lividans transformants carrying several arene dioxygenase genes. Biosci Biotechnol Biochem. 2001;65: 2472-2481. doi:10.1271/bbb.65.2472
https://doi.org/10.1271/bbb.65.2472
Saito A, Iwabuchi T, Harayama S. A novel phenanthrene dioxygenase from Nocardioides sp. Strain KP7: expression in Escherichia coli. J Bacteriol. 2000;182: 2134-2141. doi:10.1128/JB.182.8.2134-2141.2000
https://doi.org/10.1128/JB.182.8.2134-2141.2000
Wu W, Wang Z, Cong P, Li T. Accurate prediction of protein relative solvent accessibility using a balanced model. BioData Min. 2017;10: 1. doi:10.1186/s13040-016-0121-5
https://doi.org/10.1186/s13040-016-0121-5
Pathak S, Agarwal AV, Pandey VC. Phytoremediation-a holistic approach for remediation of heavy metals and metalloids. Bioremediation of Pollutants. Elsevier; 2020. pp. 3-16. doi:10.1016/B978-0-12-819025-8.00001-6
https://doi.org/10.1016/B978-0-12-819025-8.00001-6
Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30: 884-886. doi:10.1093/bioinformatics/btt607
https://doi.org/10.1093/bioinformatics/btt607
Niwa T, Ying B-W, Saito K, Jin W, Takada S, Ueda T, et al. Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc Natl Acad Sci USA. 2009;106: 4201-4206. doi:10.1073/pnas.0811922106
https://doi.org/10.1073/pnas.0811922106
Wainwright M. Dyes, trypanosomiasis and DNA: a historical and critical review. Biotech Histochem. 2010;85: 341-354. doi:10.3109/10520290903297528
https://doi.org/10.3109/10520290903297528
Samanta SK, Chakraborti AK, Jain RK. Degradation of phenanthrene by different bacteria: evidence for novel transformation sequences involving the formation of 1-naphthol. Appl Microbiol Biotechnol. 1999;53: 98-107. doi:10.1007/s002530051621
https://doi.org/10.1007/s002530051621
Tu M, Cheng S, Lu W, Du M. Advancement and prospects of bioinformatics analysis for studying bioactive peptides from food-derived protein: Sequence, structure, and functions. TrAC Trends in Analytical Chemistry. 2018;105: 7-17. doi:10.1016/j.trac.2018.04.005
https://doi.org/10.1016/j.trac.2018.04.005
Bartocci E, Lió P. Computational modeling, formal analysis, and tools for systems biology. PLoS Comput Biol. 2016;12: e1004591. doi:10.1371/journal.pcbi.1004591
Copyright (c) 2021 Authors

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.







